Creating Interactive Visualizations in R
Martin Schweinberger
2022-09-13

Introduction
This tutorial shows how to generate interactive data visualizations in R.

This tutorial is aimed at beginners and intermediate users of R with the aim of showcasing how to generate interactive visualizations and how to process the resulting concordances using R. The aim is not to provide a fully-fledged analysis but rather to show and exemplify selected useful methods associated with interactive graphs
The entire R Notebook for the tutorial can be downloaded here. If you want to render the R Notebook on your machine, i.e. knitting the document to html or a pdf, you need to make sure that you have R and RStudio installed and you also need to download the bibliography file and store it in the same folder where you store the Rmd file.
Interactive visualization refers to a type of graphic visualization that allows the viewer to interact with the data or the information that is visualized. As such, interactive visualizations are more engaging or appealing compared with non-interactive visualization. However, interactive visualizations cannot be implemented in reports that are printed on paper but are restricted to digital formats (e.g. websites, presentations, etc.).
There are various options to generate interactive data visualizations in R. The most popular option is to create a shiny app (Beeley 2013). This tutorial will not use shiny because shiny requires that the computation on which the computation that underlies the visualization is performed on a server. Rather, we will use GooleViz (Gesmann and Castillo 2011) for generating interactive visualizations that use the computer (or the browser) of the viewer to perform the computation. Thus, the interactive visualizations shown here do not require an external server.
Preparation and session set up
This tutorial is based on R. If you have not installed R or are new to it, you will find an introduction to and more information how to use R here. For this tutorials, we need to install certain packages from an R library so that the scripts shown below are executed without errors. Before turning to the code below, please install the packages by running the code below this paragraph. If you have already installed the packages mentioned below, then you can skip ahead and ignore this section. To install the necessary packages, simply run the following code - it may take some time (between 1 and 5 minutes to install all of the libraries so you do not need to worry if it takes some time).
# install packages
install.packages("googleVis")
install.packages("tidyverse")
install.packages("DT")
install.packages("flextable")
install.packages("ggplot2")
install.packages("gganimate")
install.packages("gapminder")
install.packages("maptools")
install.packages("plotly")
install.packages("leaflet")
# install klippy for copy-to-clipboard button in code chunks
install.packages("remotes")
remotes::install_github("rlesur/klippy")Now that we have installed the packages, we activate them as shown below.
# set options
options(stringsAsFactors = F) # no automatic data transformation
options("scipen" = 100, "digits" = 12) # suppress math annotation
# Warning: the following option adaptation requires re-setting during session outro!
op <- options(gvis.plot.tag='chart') # set gViz options
# activate packages
library(googleVis)
library(tidyverse)
library(DT)
library(flextable)
library(ggplot2)
library(gganimate)
library(gapminder)
library(maptools)
library(plotly)
library(leaflet)
# activate klippy for copy-to-clipboard button
klippy::klippy()Once you have installed R and RStudio and also initiated the session by executing the code shown above, you are good to go.
Getting Started
To get started with motion charts, we load the googleVis package for the visualizations, the tidyverse package for data processing, and we load a data set called coocdata. The coocdata contains information about how often adjectives were amplified by a degree adverb across time (see below).
# load data
coocdata <- base::readRDS(url("https://slcladal.github.io/data/coo.rda", "rb"))Decade | Amp | Adjective | N.Amp. | N.Adj. | OBS | EXP | Probability | CollStrength | OddsRatio | p | attr | sig |
1,870 | pretty | amiable | 7 | 4 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | amusing | 7 | 4 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | angry | 7 | 8 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | annoyed | 7 | 1 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | bad | 7 | 7 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | beautiful | 7 | 16 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | busy | 7 | 108 | 0 | 0.1 | 0.02 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | charming | 7 | 7 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | clever | 7 | 4 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | comfortable | 7 | 7 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | contemptible | 7 | 2 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | convenient | 7 | 13 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | dangerous | 7 | 44 | 0 | 0.1 | 0.01 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | dark | 7 | 11 | 0 | 0.0 | 0.00 | 0 | 0 | 1 | repel | n.s. |
1,870 | pretty | different | 7 | 202 | 0 | 0.2 | 0.03 | 0 | 0 | 1 | repel | n.s. |
The coocdata is rather complex and requires some processing. First, we rename the columns to render their naming more meaningful. In this context we rename the OBS column Frequency and the Amp column Amplifier. As we are only interested if an adjective was amplified by very, we collapse all amplifiers that are not very in a bin category called other. We then calculate the frequency of the adjective within each time period and also the frequency with which each adjective is amplified by either very or other amplifiers. Then, we calculate the percentage with which each adjective is amplified by very.
# process data
coocs <- coocdata %>%
dplyr::select(Decade, Amp, Adjective, OBS) %>%
dplyr::rename(Frequency = OBS,
Amplifier = Amp) %>%
dplyr::mutate(Amplifier = ifelse(Amplifier == "very", "very", "other")) %>%
dplyr::group_by(Decade, Adjective, Amplifier) %>%
dplyr::summarise(Frequency = sum(Frequency)) %>%
dplyr::ungroup() %>%
tidyr::spread(Amplifier, Frequency) %>%
dplyr::group_by(Decade, Adjective) %>%
dplyr::mutate(Frequency_Adjective = sum(other + very),
Percent_very = round(very/(other+very)*100, 2)) %>%
dplyr::mutate(Percent_very = ifelse(is.na(Percent_very), 0, Percent_very),
Adjective = factor(Adjective))Decade | Adjective | other | very | Frequency_Adjective | Percent_very |
1,870 | amiable | 0 | 0 | 0 | 0 |
1,870 | amusing | 0 | 0 | 0 | 0 |
1,870 | angry | 0 | 0 | 0 | 0 |
1,870 | annoyed | 0 | 0 | 0 | 0 |
1,870 | bad | 0 | 0 | 0 | 0 |
1,870 | beautiful | 0 | 0 | 0 | 0 |
1,870 | busy | 1 | 0 | 1 | 0 |
1,870 | charming | 0 | 0 | 0 | 0 |
1,870 | clever | 0 | 0 | 0 | 0 |
1,870 | comfortable | 0 | 0 | 0 | 0 |
We now have a data set that we can use to generate interactive visualization.
1 Basic Interactive Graphs
This section shows some very basic interactive graphs including scatter plots, line graphs, and bar plots.
Scatter Plots
Scatter plots show the relationship between two numeric variables if you have more than one observation per variable level (if the data is not grouped by another variable). This means that you can use scatter plots to display data when you have, e.g. more than one observation for each data in your data set. If you only have a single observation, you could also use a line graph (which we will turn to below).
scdat <- coocs %>%
dplyr::group_by(Decade) %>%
dplyr::summarise(Precent_very = mean(Percent_very))
# create scatter plot
SC <- gvisScatterChart(scdat,
options=list(
title="Interactive Scatter Plot",
legend="none",
pointSize=5))If you want to display the visualization in a Notebook environment, you can use the plot function as shown below.
plot(SC)However, if you want to display the visualization on a website, you must use the print function rather than the plot function and specify that you want to print a chart.
print(SC, 'chart')Line Graphs
To create an interactive line chart, we use the gvisLineChart function as shown below.
# create scatter plot
SC <- gvisLineChart(scdat,
options=list(
title="Interactive Scatter Plot",
legend="none"))If you want to display the visualization in a Notebook environment, you can use the plot function. For website, you must use the print function and specify that you want to print a chart.
print(SC, 'chart')Bar Plots
To create an interactive bar chart, we use the gvisBarChart function as shown below.
# create scatter plot
SC <- gvisBarChart(scdat,
options=list(
title="Interactive Bar chart",
legend="right",
pointSize=10))Normally, you would use the plot function to display the interactive chart but you must use the print function with the chart argument if you want to display the result on a website.
print(SC, 'chart')2 Animations
Animations or GIFs (Graphics Interchange Format) can be generated using the gganimate package written by Thomas Lin Pedersen and David Robinson. The gganimate package allows to track changes over time while simultaneously displaying several variables in one visualization. As we will create animations using the ggplot2 package, we also load that package from the library. In this case, we use the gapminder data set which comes with the gapminder package and which contains information about different countries, such as the average life expectancy, the population, or the gross domestic product (GDP), across time.
# set options
theme_set(theme_bw())country | continent | year | lifeExp | pop | gdpPercap |
Afghanistan | Asia | 1,952 | 28.801 | 8,425,333 | 779.4453145 |
Afghanistan | Asia | 1,957 | 30.332 | 9,240,934 | 820.8530296 |
Afghanistan | Asia | 1,962 | 31.997 | 10,267,083 | 853.1007100 |
Afghanistan | Asia | 1,967 | 34.020 | 11,537,966 | 836.1971382 |
Afghanistan | Asia | 1,972 | 36.088 | 13,079,460 | 739.9811058 |
Afghanistan | Asia | 1,977 | 38.438 | 14,880,372 | 786.1133600 |
Afghanistan | Asia | 1,982 | 39.854 | 12,881,816 | 978.0114388 |
Afghanistan | Asia | 1,987 | 40.822 | 13,867,957 | 852.3959448 |
Afghanistan | Asia | 1,992 | 41.674 | 16,317,921 | 649.3413952 |
Afghanistan | Asia | 1,997 | 41.763 | 22,227,415 | 635.3413510 |
After loading the data, we create static plot so that we can check what the data looks like at one point in time.
p <- ggplot(
gapminder,
aes(x = gdpPercap, y=lifeExp, size = pop, colour = country)
) +
geom_point(show.legend = FALSE, alpha = 0.7) +
scale_color_viridis_d() +
scale_size(range = c(2, 12)) +
scale_x_log10() +
labs(x = "GDP per capita", y = "Life expectancy")
p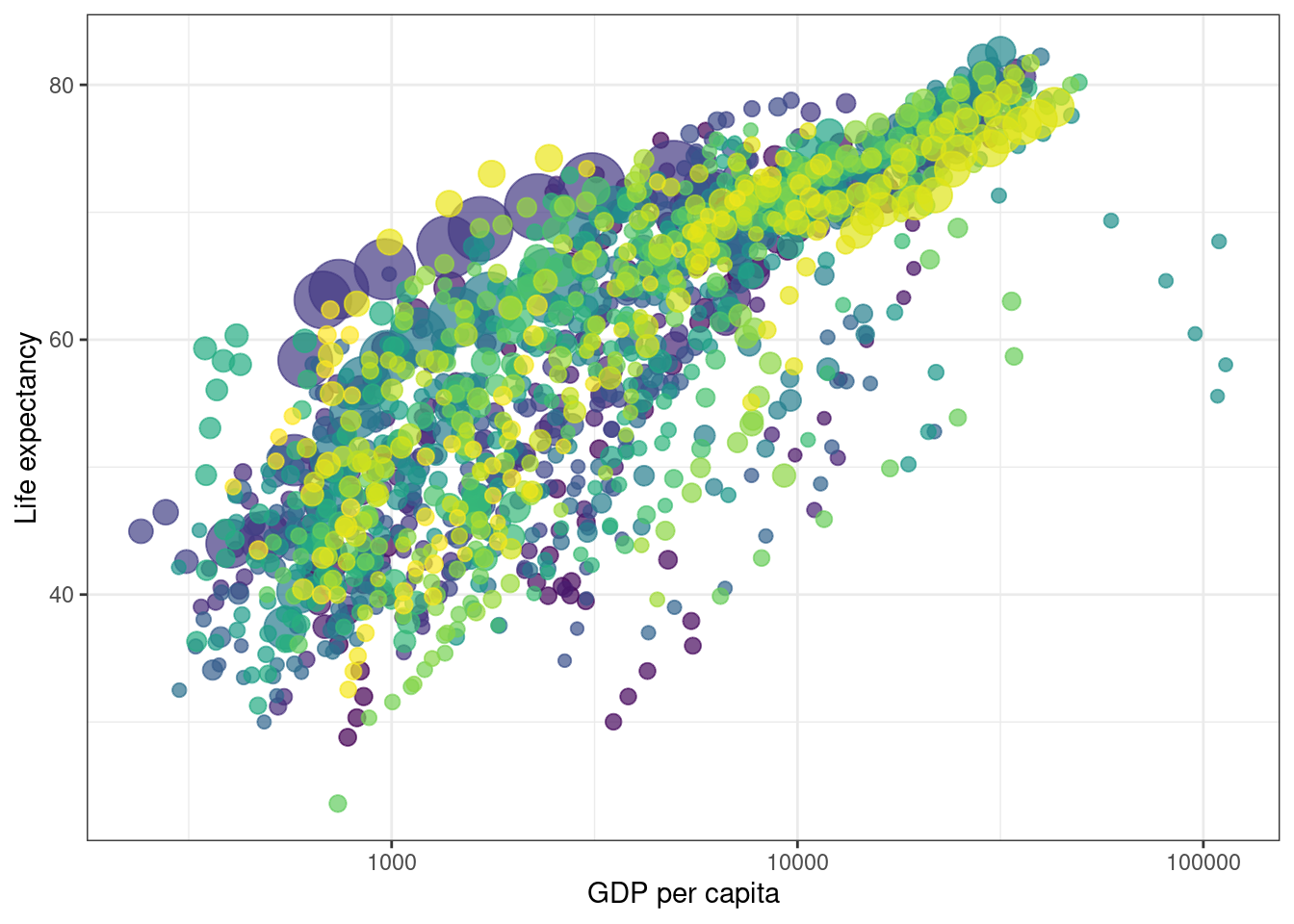
We can then turn static plot into animation by defining the content of the transition_time object.
gif <- p + transition_time(year) +
labs(title = "Year: {frame_time}")
# show gif
gif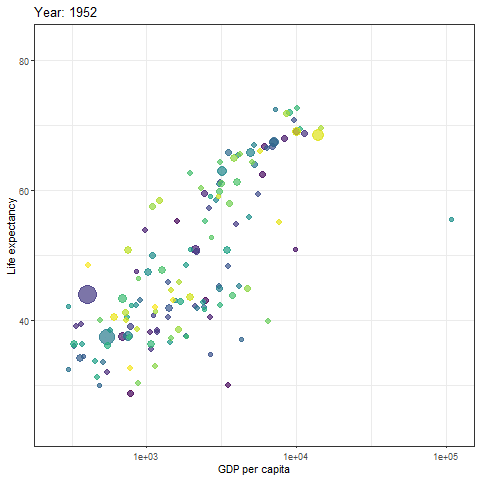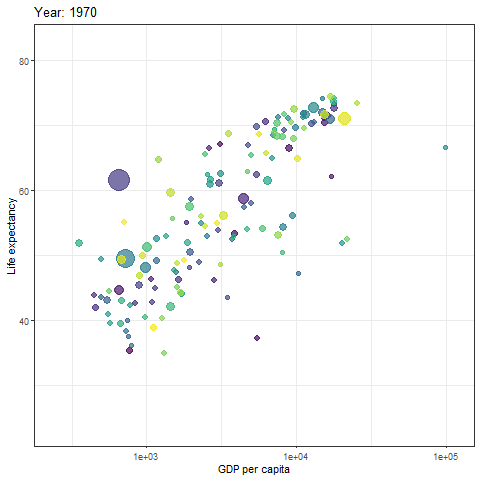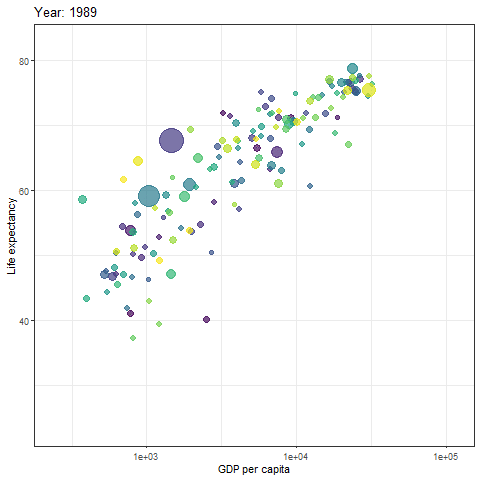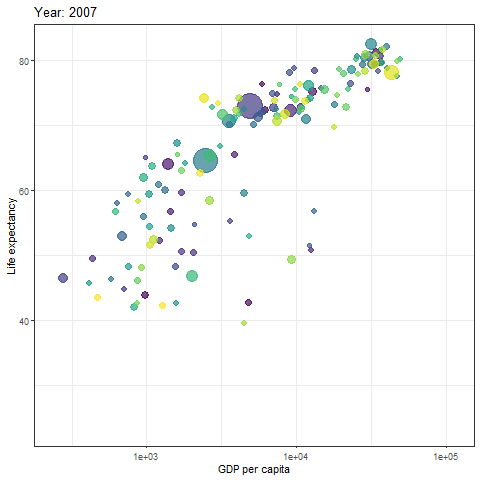
Another way to generate animations is to use the plotly package as shown below. While I personally do not find the visualizations created by the plot_ly function as visually appealing, it has the advantage that it allows mouse-over effects.
fig <- gapminder %>%
plot_ly(
x = ~gdpPercap,
y = ~lifeExp,
size = ~pop,
color = ~continent,
frame = ~year,
text = ~country,
hoverinfo = "text",
type = 'scatter',
mode = 'markers'
)
fig <- fig %>% layout(
xaxis = list(
type = "log"
)
)
fig3 Interactive Maps
You can also use the leaflet package to create interactive maps. In this example, we display the beautiful city of Brisbane and the visualization allows you to zoom in and out.
# generate visualization
m <- leaflet() %>%
setView(lng = 153.05, lat = -27.45, zoom = 12)
# display map
m %>% addTiles()Another option is to display information about different countries. In this case, we can use the information provided in the maptools package which comes with a SpatialPolygonsDataFrame of the world and the population by country (in 2005). To make the visualization a bit more appealing, we will calculate the population density, add this variable to the data which underlies the visualization, and then display the information interactively. In this case, this means that you can use mouse-over or hoover effects so that you see the population density in each country if you put the cursor on that country (given the information is available for that country).
We start by loading the required package from the library, adding population density to the data, and removing data points without meaningful information (e.g. we set values like Inf to NA).
# load data
data(wrld_simpl)
# calculate population density and add it to the data
wrld_simpl@data$PopulationDensity <- round(wrld_simpl@data$POP2005/wrld_simpl@data$AREA,2)
wrld_simpl@data$PopulationDensity <- ifelse(wrld_simpl@data$PopulationDensity == "Inf", NA, wrld_simpl@data$PopulationDensity)
wrld_simpl@data$PopulationDensity <- ifelse(wrld_simpl@data$PopulationDensity == "NaN", NA, wrld_simpl@data$PopulationDensity)
# inspect data
head(wrld_simpl@data)## FIPS ISO2 ISO3 UN NAME AREA POP2005 REGION SUBREGION
## ATG AC AG ATG 28 Antigua and Barbuda 44 83039 19 29
## DZA AG DZ DZA 12 Algeria 238174 32854159 2 15
## AZE AJ AZ AZE 31 Azerbaijan 8260 8352021 142 145
## ALB AL AL ALB 8 Albania 2740 3153731 150 39
## ARM AM AM ARM 51 Armenia 2820 3017661 142 145
## AGO AO AO AGO 24 Angola 124670 16095214 2 17
## LON LAT PopulationDensity
## ATG -61.783 17.078 1887.25
## DZA 2.632 28.163 137.94
## AZE 47.395 40.430 1011.14
## ALB 20.068 41.143 1151.00
## ARM 44.563 40.534 1070.09
## AGO 17.544 -12.296 129.10We can now display the data.
# define colors
qpal <- colorQuantile(rev(viridis::viridis(10)),
wrld_simpl$PopulationDensity, n=10)
# generate visualization
l <- leaflet(wrld_simpl, options =
leafletOptions(attributionControl = FALSE, minzoom=1.5)) %>%
addPolygons(
label=~stringr::str_c(
NAME, ' ',
formatC(PopulationDensity, big.mark = ',', format='d')),
labelOptions= labelOptions(direction = 'auto'),
weight=1, color='#333333', opacity=1,
fillColor = ~qpal(PopulationDensity), fillOpacity = 1,
highlightOptions = highlightOptions(
color='#000000', weight = 2,
bringToFront = TRUE, sendToBack = TRUE)
) %>%
addLegend(
"topright", pal = qpal, values = ~PopulationDensity,
title = htmltools::HTML("Population density <br> (2005)"),
opacity = 1 )
# display visualization
lSession Outro
When generating interactive maps, it is important that you reset the default graphics parameters which had to be adapted in the session set-up. Therefore, in a final step, we restore the default graphics options.
## Set options back to original options
options(op)Citation & Session Info
Schweinberger, Martin. 2022. Creating Interactive Visualization in R. Brisbane: The University of Queensland. url: https://slcladal.github.io/motion.html (Version 2022.09.13)
@manual{schweinberger2021motion,
author = {Schweinberger, Martin},
title = {Creating motion charts using R},
note = {https://slcladal.github.io/motion.html},
year = {2022},
organization = "The University of Queensland, Australia. School of Languages and Cultures},
address = {Brisbane},
edition = {2022.09.13}
}sessionInfo()## R version 4.2.1 (2022-06-23)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 22.04.1 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
##
## locale:
## [1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
## [5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
## [7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices datasets utils methods base
##
## other attached packages:
## [1] leaflet_2.1.1 plotly_4.10.0 maptools_1.1-4 sp_1.5-0
## [5] gapminder_0.3.0 gganimate_1.0.7 flextable_0.7.3 DT_0.24
## [9] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.9 purrr_0.3.4
## [13] readr_2.1.2 tidyr_1.2.0 tibble_3.1.7 ggplot2_3.3.6
## [17] tidyverse_1.3.2 googleVis_0.7.0
##
## loaded via a namespace (and not attached):
## [1] fs_1.5.2 lubridate_1.8.0 RColorBrewer_1.1-3
## [4] progress_1.2.2 httr_1.4.3 tools_4.2.1
## [7] backports_1.4.1 bslib_0.3.1 utf8_1.2.2
## [10] R6_2.5.1 lazyeval_0.2.2 DBI_1.1.3
## [13] colorspace_2.0-3 withr_2.5.0 gridExtra_2.3
## [16] tidyselect_1.1.2 prettyunits_1.1.1 compiler_4.2.1
## [19] cli_3.3.0 rvest_1.0.2 xml2_1.3.3
## [22] officer_0.4.3 labeling_0.4.2 sass_0.4.1
## [25] scales_1.2.0 systemfonts_1.0.4 digest_0.6.29
## [28] foreign_0.8-82 rmarkdown_2.14 base64enc_0.1-3
## [31] pkgconfig_2.0.3 htmltools_0.5.2 dbplyr_2.2.1
## [34] fastmap_1.1.0 highr_0.9 htmlwidgets_1.5.4
## [37] rlang_1.0.4 readxl_1.4.0 jquerylib_0.1.4
## [40] generics_0.1.3 farver_2.1.1 jsonlite_1.8.0
## [43] crosstalk_1.2.0 zip_2.2.0 googlesheets4_1.0.0
## [46] magrittr_2.0.3 Rcpp_1.0.8.3 munsell_0.5.0
## [49] fansi_1.0.3 viridis_0.6.2 gdtools_0.2.4
## [52] lifecycle_1.0.1 stringi_1.7.8 yaml_2.3.5
## [55] grid_4.2.1 crayon_1.5.1 lattice_0.20-45
## [58] haven_2.5.0 hms_1.1.1 magick_2.7.3
## [61] klippy_0.0.0.9500 knitr_1.39 pillar_1.7.0
## [64] uuid_1.1-0 codetools_0.2-18 reprex_2.0.1
## [67] glue_1.6.2 evaluate_0.15 data.table_1.14.2
## [70] renv_0.15.4 modelr_0.1.8 vctrs_0.4.1
## [73] tzdb_0.3.0 tweenr_1.0.2 cellranger_1.1.0
## [76] gtable_0.3.0 assertthat_0.2.1 xfun_0.31
## [79] broom_1.0.0 viridisLite_0.4.0 googledrive_2.0.0
## [82] gargle_1.2.0 ellipsis_0.3.2References
Beeley, Chris. 2013. Web Application Development with R Using Shiny. Packt Publishing Ltd.
Gesmann, Markus, and Diego de Castillo. 2011. “Using the Google Visualisation Api with R.” The R Journal 3 (2): 40–44.